Neuroscience Notes - Molecular Biology
Some notes taken from Neuroscience course, part 1: Molecular Biology.
last update: Sep. 16, 2020
Translation of Terms
| Eng | Chs | Comment |
|---|---|---|
| nucleus | 细胞核 | |
| chromosome | 染色体 | |
| polymerase | 聚合酶 | |
| cytoplasm | 细胞质 | |
| ribosome | 核糖体 | |
| polymer | 聚合物 | |
| deoxyribonucleotide | 脱氧核糖核酸 | DNA |
| nucleotide | 核酸 | |
| nomenclature | 命名法/术语 | 3’ 5’末端 |
| phosphate | 磷酸盐 | |
| hydroxyl | 羟基 | -OH |
| plasmid | 质粒 | |
| ATP | 三磷酸腺苷 | adenosine triphosphate |
| phosphodiester | 磷酸二酯 | breaking ~ bond -> energy |
| polypeptide | 多肽 | |
| codon | 密码子 | |
| protease | 蛋白酶 | |
| catalytic | 催化的 | |
| denaturation | 使变性 | (化学意义上 |
| Hybridization | 杂交? | 两条相配的DNA(RNA)链 |
| phage | 噬菌体 | |
| molar | 摩尔的 | |
| oligonucleotide | 低聚核苷酸 | short (DNAs) |
| triphosphate | 三磷酸盐 | |
| southern/northern blot | 南/北方墨点法 | |
| thyrotropin | 促甲状腺素 | |
| glutamate | 谷氨酸 | |
| retroviruse | 逆转录病毒 | |
| agar | 琼脂 | 培养基材料 |
| E. Coli | 大肠杆菌 | |
| mammalian | 哺乳类动物的 | |
| eukaryotic | 真核的 | eukaryote 真核细胞 |
| endonuclease | 内切酶 | DNA clone |
| ligase | 连接酶 | DNA clone |
| primer | 引物 | e.g., in PCR |
| acrylamide | 丙烯酰胺 | used in chemical DNA sequencing |
| dideoxy | 双脱氧法 | used in sequencing |
| pyrophosphate | 焦磷酸盐 | used in sequencing |
| eppendorf tube | 微量离心管 | |
| hydrophobic | 疏水的 | |
| exon | 外显子 | |
| centromere | 着丝粒 | |
| embryonic | 胚胎的 | |
| pluripotent | 多功能的 | |
| neomycin | 新霉素 | |
| cell culture | 细胞培养 | |
| embryonic stem cell | 胚胎干细胞 | |
| formaldehyde | 甲醛 | |
| chromophore | 发色团 | |
| acetylcholine | 乙酰胆碱 | a neurotransmitter |
| cholinergic | 胆碱能的 | |
| bacteriophage | 噬菌体 | |
| recombinase | 重组酶 | |
| cardiac | 心脏的 | |
| phenotype | 表现型 | [遗传] |
| serotonin | 血清素 | 神经递质 |
| estrogen | 雄性激素 | |
| steroid | 类固醇 | |
| axonal | 轴突的 | |
| lipid | 脂类 | |
| nucleosome | 核小体 | |
| histone | 组蛋白 | |
| epigenetic | 表观遗传的 | |
| polyadenylation | 聚腺苷酸化 | |
| kinase | 激酶;致活酶 | |
| dimer | 二聚体 | |
| ligand | 配位体 | |
| acetylation | 乙酰化作用 | |
| in vitro | 在体外 | |
| corpus callosum | 胼胝体 | |
| dendrite | 树突 | |
| interphase | 间期 | 不再分裂的细胞阶段 |
| halorhodopsin | 鹽細菌視紫紅質 | |
| hypocretin | 下视丘分泌素 | 食欲素,和失眠也有关 |
| narcolepsy | 嗜睡症 | |
| dopamine | 多巴胺 | |
| ligand | 配位体 | |
| pharmacological | 药理学的 |
Lecture
9.1
- intron & exon
- primer design
DNA and RNA
感觉一节课把我整个高中学过的东西都讲完了。
- DNA strand: a sugar/phosphate backbone; polarity of (5’ to 3’)
- DNA can be double stranded ds or single stranded ss, stability
- “Wastson-Crick” double helical conformation, A=(2H)=T, C=(3H)=G
- Increase temperature or change pH(>12.5): ds -> ss
- DNA can be linear (humnan) or circular (bacterial plasmid)**
- ds: oriented in an antiparallel configuration
- sense strand: seq from cDNA (DNA copy of an mRNA)
- Polymerize: dATP => DNA; rATP => RNA
- Synthesis always occurs from 5’ to 3’: Leading and Lagging (discontinuous) strand.
- RNA is susceptible to degradation by nucleases
- DNAs & RNAs can be size-separated using gel electrophoresis: negatively charged
References



Central Dogma 中央教条中心法则
- DNA —(Transcription)–> RNA —(Translation)–> Protein
| General | Special | Unknown |
|---|---|---|
| DNA → DNA | RNA → DNA | protein → DNA |
| DNA → RNA | RNA → RNA | protein → RNA |
| RNA → protein | DNA → protein | protein → protein |
DNA Hybridization
- ds DNA can find their matching partners even after cooling from ss DNAs
- DNA “melting” ds -> ss
- After cooling down, ss DNAs hybridize
Factors => Melting Temperature $T_m$
- Salt concentration affect melting temp. Higher salt -> higher temp
- GC pair more stable because: G≡C, A=T
- DNA length: longer -> higher temp
Factors => Hybridization
- Time of half of the molecules to reassociate $t_{1/2}$
- Inversely proportional to the molarity of those molecules
- Also influenced by temp, salt concentration, DNA length
- Optimal temp: ~25°C below the $T_m$
- Short DNAs (oligonucleotides, primers) => larger targets
- Approximate formula: $t_{1/2} = 1 day/[DNA]$, [DNA] in picomolar(PM)
Applications
- PCR (“polymerase chain reaction”)
公主链接 - Production of cDNA
- DNA sequencing
- Immobilize nucleic acids
- Hybridization-mediated: RNA interference & CRISPR]
Cloning DNAs
- Clone DNA copies from mRNAs <= reverse transcriptase enzyme
- cDNA was originally to study how each gene product functioned
Reverse Transcriptase
Reverse transcriptase is a primer-dependent polymerase that will synthesize DNA from the RNA template.
- Convert DNA into a ds DNA
- Analyzing DNAs is easier than RNAs
- DNA oligonucleotides are used to “prime” first strand cDNA synthesis.
- Using random primers/oligo-d(T) as a primer
Cloning
The first technique that provided an abundant source of a specific DNA
- Foreign DNA + Cloning vector are cut by the same Restriction Endonucleases
- They are then Covalently joined by DNA ligase
- Put the Cloned DNA vector into E.Coli
- Most frequently used cloning vector: a small circular double-stranded DNA called a “plasmid”
- Cloning involves inserting a specific DNA fragment into a vector and propagating the recombinant plasmid
- Each bacterium should only contain a single recombinant plasmid
- DNAs can be cleaved with restriction endonucleases recognizing specific nucleotide sequences and resulting in “sticky ends”
- E.g. EcoR1: GAATTC => G + AATTC & CTTAA + G
Plasmids
Plasmids are (small) extrachromosomal circular double-stranded DNAs that replicate independently from the host chromosome (E. coli).
Plasmids used for cloning contain an origin of replication (“replicon”), selectable marker >(typically antibiotic resistance) and specific sites into which the DNA fragment can be inserted (a “multiple cloning site” or “polylinker”).
- Selectable markers typically (but not exclusively) a gene conferring antibiotic resistance. Two examples are ampicillin (“aminopenicillin”) resistance (AmpR) and kanamycin resistance (KanR).
- Replicon: (Origin of replication and control elements) This is the region of the plasmid that is required for replication. Many commonly used plasmids have a “pUC” type origin permitting replication in E. Coli.
- Ampicillin inhibits bacterial cell wall synthesis, greatly slowing bacterial growth. The AmpR gene encodes beta-lactamase, an enzyme that cleaves a ring in the structure of ampicillin, thereby inactivating it.
- Kanamycin interacts with multiple ribosomal proteins and inhibits protein synthesis. The KanR gene encodes an aminophosphotransferase that phosphorylates kanamycin, preventing it from binding to the ribosomal proteins.
- Multiple cloning site (MCS or polylinker) This region contains multiple, adjacent “unique” sites of restriction endonuclease cleavage– providing many ways to introduce our “inserts”.
Subcloning
- Molecular cloning refers to the insertion of a DNA fragment into a plasmid
- Subcloning: to “move” DNAs from one plasmid to another, in order to obtain expression tailored to their needs.
PCR
Polymerase Chain Reaction 聚合酶链式反应

- Enabling the mass production of DNA without to clone it
- Exponential amplification of DNA, doubling DNA mass with every cycle
Process
PCR requires: 2 opposing small DNA primers
- Step 1, 2, 3 represents 3 thermal cycles
- Denaturation: separate 2 strands via heating
- Annealing: reduce temp to permit the hybridization of primers
- Extension: raise temp to the optimal temp for heat-stable polymerase to synthesis DNA
- DNA fragments doubled with each cycle
- Hybridization of short DNA => to target/template molecule
- Primers define the boundaries of the amplified region (5’ -> 3’)
- But we can also add additional nucleotides to the primers (e.g. sticky ends)
- Heat-stable enzyme: Taq(generate sticky ends), Pfu&Q5(don’t)
- High Fidelity enzyme? fewer mistakes
- Designing PCR primers: need both upstream/sense and downstream/antisense
- Can also targeting a specific region (Open reading frame)
DNA Sequencing
A target of cloning is sequencing.
- Cycle sequencing
- Capillary sequencing
Development in Bio Technologies
- Gene cloning
- DNA sequencing
- PCR
- RNA interference
- DNA sequencing (NG)
- CRISPR
Chemical Sequencing
- Chemical modification Use different chemical modifications of the DNA that permit the subsequent specific cleavage of these molecules: at G, G/A, T/C, C
- Different-sized DNAs Resulted in the generation of a distribution of different-sized DNAs
- Radioactively labeled DNAs were “end-labeled” using T4 polynucleotide kinase and a radiolabeled form of ATP (“gamma-32P-ATP”).
Sanger/Dideoxy Sequencing
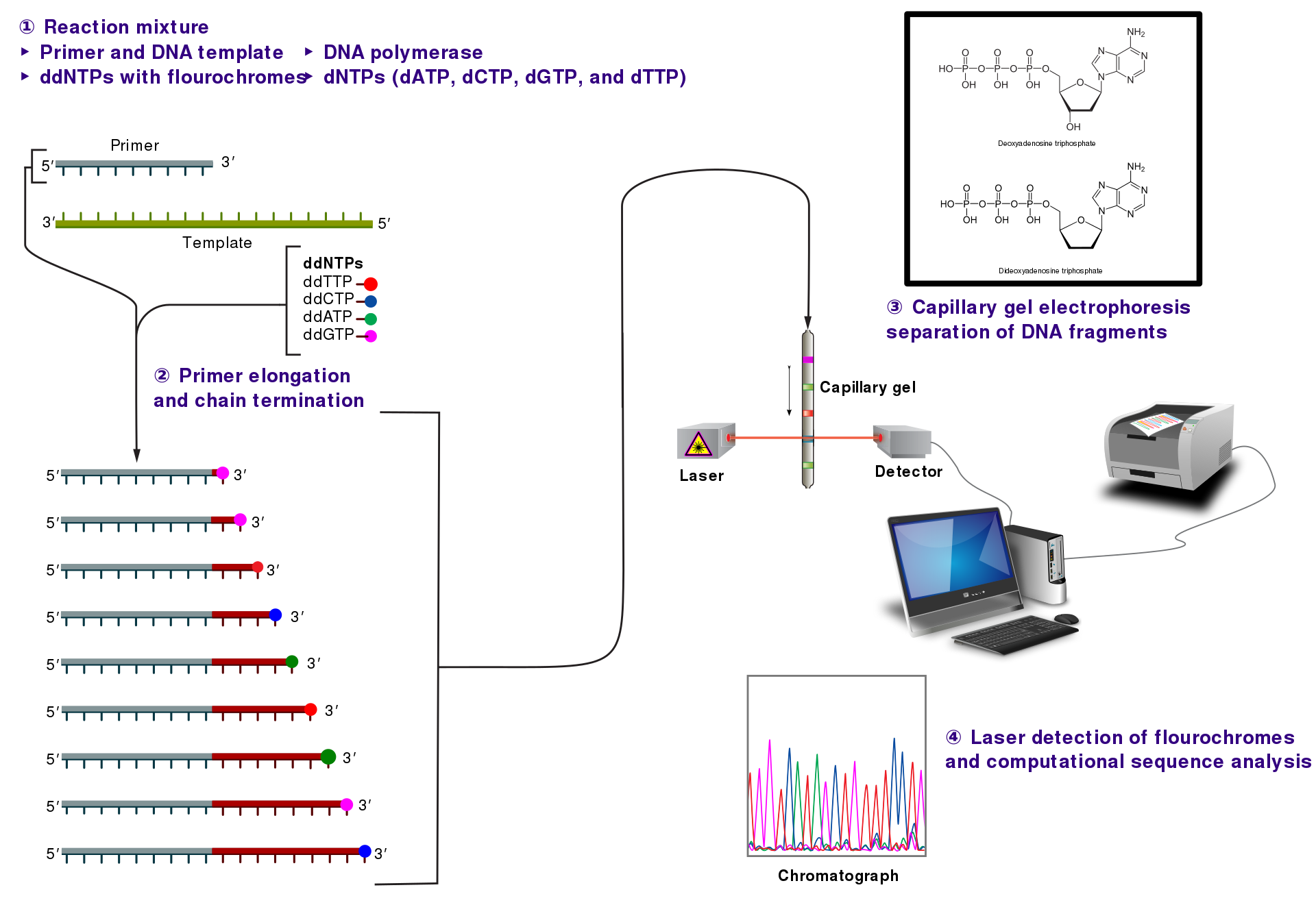
- Dideoxynucleotide(ddNTP): X 3’ -OH => termination of growing DNA strand
- In the original format, each tube contained all 4 dNTPs but just one ddNTP
- The chain-termination happened infrequently => a broad rage of sized-DNAs
- All the DNAs are “single-stranded” and all must share
Fluorescently Tagged Primers
The 4 different “colors”, permitted loading the sample onto a single lane.
“Shotgun” Sequencing
For very long sequences: human, mouse, etc.
- DNA samples is first broken into small pieces
- The data is assembled bioinformatically <= overlapping information
NG Sequencing
- High Throughput
- Require Imaging
- Detect the release of “pyrophosphate” or “H+”
Technologies
- 454 (Roche) “pyrosequencing”
- Illumina/Solexa sequencing (short seq, color of light)
- Ion semiconductor sequencing (“Ion torrent”), monitoring the release of H+
- Single molecule “real-time” (“SMRT”) sequencing (long seq)
- “Nanopore” sequencing
Human Genome Project
There is no ideal human genome reference sequence as we are all different
- Protein-coding genes: ~20k
- Non-coding genes: encode RNAs are never translated into proteins
- Alternative splicing: a mature mRNA need not include every exon present within a gene => increases the number of proteins can be made from one gene
- Organismal complexity may be related to differences in gene regulation.
- Reference Sequence: Not complete => “NGS” to sequence individuals
- SNP: single nucleotide polymorphisms ~3.5M
- CNV: copy number variants, ~1000 large (>500 bp)
- Genome Projects of different populations and higher coverage
What Genomics Make Human Human?
- Humans and chimps were ~98.77% identical in shared regions
- FOXP2 had been identified as the gene mutated in a British family having an inherited speech disorder
- FOXP2 plays a role in “vocalization” but it is not the “language” gene separating apes from man
- The human brain is ~3 times larger than that of the chimp
- Overexpression of NOTCH2NL in human neural cortical progenitors in vitro leads to an increase in cell number
Precision Medicine
- to predict who is at risk
- to diagnose disorders
- permit “personalized” health care
- permit the rapid diagnosis of pathogens
- ethical questions: privacy & disclosure
RNA Sequencing
- RNAs are not being directly sequenced but are converted to DNAs using reverse transcriptase
- Single cell RNA seq: Linnarsson lab neural cell profiling
- Cells can be categorized by similarities in their gene expression profiles using RNAseq
Transgenic & Knockout Mice
- Transgenic mice contain modified DNAs that have “randomly integrated” into the genome
- Knockout mice have been traditionally made by using homologous recombination.
- The normal allele is modified via replacement with an altered DNA that eliminates the expression of that gene
Transgenic
May insert transgenes into ant chromosome.
- The DNA construct used for the production of a typical transgenic mouse contains a promoter, protein-coding region, poly (A) addition site and an intron.
- the inserted DNAs are frequently found in tandem arrays, i.e. there are multiple adjacent copies of each transgene.
Knockout mice
- Actual recombination occurs in embryonic stem cells (“ES cells”) and not in the fertiilized egg
- “targeting construct”: selectable marker()neomycin resistance + associated elements flanked by “homology arms”
- The arms are regions of DNA that permit precise homologous recombination into the normal gene locus.
- The recombination event is designed to eliminate one or more of the gene’s exons.
- A second marker (HSV-tk) is used to select against the random insertion of the construct.
- A few of these cells are injected into early stage pre-implantation embryos (blastocysts)
- It is only useful if the altered gene is “in the germline”

Fluorescent Proteins
GFP fluorescence occurs when aequorin interacts with Ca++, which emits blue light. Some of this is transferred to GFP in a non-radiative manner, which then emits in the green part of the spectrum.
- to “tag” proteins permitted visualizing those proteins in living cells
- Variants: ECFP, EGFP, EYFP, mCherry
- The excitation energy must be higher (shorter wavelength) than that of the emitted light!!!
- BACs(Bacterial artificial chromosomes) provide a source of “promoters”
Usage
- One can label specific cell types in an organism, which typically means marking the expression of cells that express a specific promoter
- Protein function may be compromised by the presence of the tag.
- Fluorescent proteins are useful for studying the expression/localization or “co-localization” or proteins
- As a proximity detector between 2 fluorescent protein-tagged molecules.
- Collect electron microscopy data from each section.
Cre-loxP Recombination
Cre recombinase is an enzyme derived from a bacterial virus that mediates recombination when it detects its target sequence (a “loxP” site).
- Two recombinases: Cre(bacteriophage P1) and Flp(yeast)
- Cre: recognizes a 34 bp sequence (“loxP”) that contains an inverted repeat
- Recombination requires 2 sites, and each has a direction => excise or flip
- Knockout mice are generated by homologous recombination in embryonic stem cells
- Conditional knockout mice: contain exons flanked by loxP sites. When mated to cre-expressing lines, the doubly transgenic offspring should undergo a loss of the flanked exon in the cell types that express cre recombinase.
- Two lines: Cre driver line and Floxed line
- lox-STOP-lox cassette (transcriptional stop)
- There are different kinds of “mutant” loxP sites and they do not work with the other loxP sites.
- “intersectional” strategies(“binary systems”): limit manipulations to defined sets of cells
- Temporal regulation: steroid inducible cre (“Cre-ERT2”). In the presence of estrogen (or tamoxifen), the fusion protein enters the nucleus=> permits one to regulate the timing of the induction of cre activity.
Brainbow mice
- The Brainbow mice: express mixtures of fluorescent proteins
- The Brainbow mice express multiple (5 or more) tandem copies of their transgenes => each cell can express a different fluorescent protein from each copy of the transgene
- Goal: understand connections; limited successes in the mammalian brain because of its complexity
- Brainbow version 3: these proteins are farnesylated (a lipid modification), which permits membrane association => detect fine axonal processes
CLARITY
- The fixed tissues are opaque (perfuses” the tissue to eliminate the blood and to also introduce a fixative to preserve the tissue morphology)
- CLARITY: a brain is perfused with a mixture of acrylamides, formaldehyde and a cross-linking agent => The lipids (which contribute to brain opacity) are removed via electrophoresis => clear for imaging
3D “EM” Reconstruction
Kasthuri et al., 2015, Cell 162:648-661.
- Determine the synaptic connectivity of the mammalian brain.
- Murine brain tissue fixed for electron microscopy is sectioned and transferred onto a moving plastic tape
- The tape is cut and placed onto a silicon support.
- These are inserted into a specially designed electron microscope.
- Adjacent sections need to be connected to form a 3D image of a region of tissue
- Typically only one out of nine axon actually formed a synapse.
Regulation of Gene Expression
The regulation of gene expression: Genomic organization
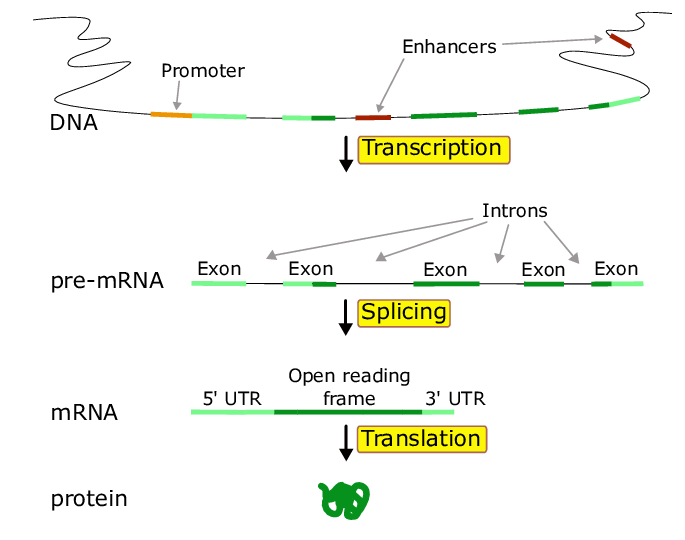
- Promoter: the region near the 5’ end of a gene that contains the cis-acting regulatory elements that bind to transcription factors => that facilitate the ability of RNA polymerase to bind and initiate “transcription”
- Enhancers: (distant sites) regions of DNA that regulate a gene’s expression
- Regulatory elementsL protein fatcors (trans elements) and DNA sequences (cis elements)
Transcription
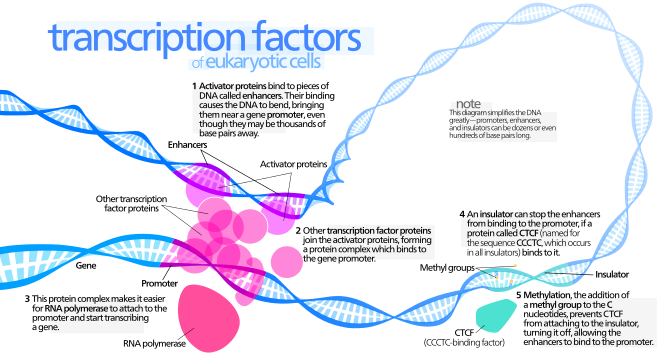

- Transcription factors: proteins that can bind to DNA(enhancers & promter) & promote transcription.
- Transcription factor binding to the promoter and to enhancers is thought to lead to the recruitment of the large collection of molecules required to initiate the production of RNA by RNA polymerase (to initiate transcription).
- Each interphase chromosome is largely confined to a “territory” within the nucleus.
- RNA Polymerase: I: ribosomal RNA; II: mRNA/snRNA/miRNA; III: small RNAs (tRNA, etc.)
- One surprise: Transcription not only occurs at promoters but can also be initiated at enhancers and the these transcripts are called “eRNAs”.
Topological Associating Domains (TADs)
In interphase chromosomes, the chromatin is thought to be organized into loops that are ~1 Mb in length. The loops (or collections of loops) are called “topological associating domains” (TADs), which contain multiple genes.
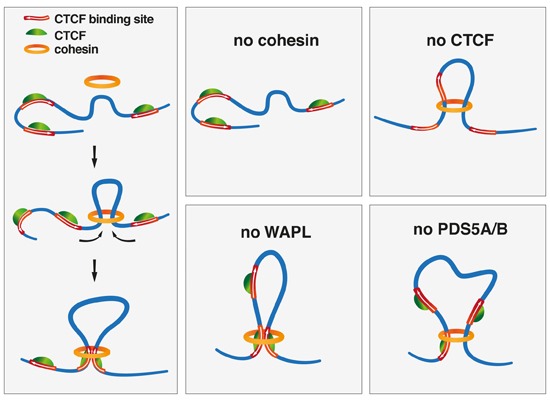
Ref:TAD
- chromosome is organized into “sub-regions” called TADs
- to identify if non-adjacent sites on chromosomes could interact
- chromosomes are divided into distinct regulatory “neighborhoods”, adjacent TADs are thought to be insulated from each other
- TADs solution: restricting the activity of enhancers to specific promoters
- Chromosome conformation capture (3C): determine the “loop structure”
- cohesin complex: extrude DNA; the presence of CTCF appears to terminate loop formation
- TADs <= one or more loops; loop boundaries <= CTCF & cohesin
- chromosome structure is dynamic and that TAD boundaries shift
- loop structure may not be the primary feature guiding gene regulation
Stages
- Initiation: RNA polymerase and other factors are assembled into a complex (the pre-initiation complex) that permits transcription to start
- “pre-initiation complex” (PIC
position-independent code): general trascription factors & “the mediator complex” - unwinds the promoter DNA (TFIIH) => RNA pol gain access to DNA template
- cell-type sepcific differneces in this complex
- “pre-initiation complex” (PIC
- Pausing: Shortly after initiation, RNA Pol II pauses and its ability to proceed is a regulated step. => ability to rapidly increase transcription
- transcribes 50~80 NTs before pausing
- Elongation Once the pause has been lifted, RNA polymerase moves down the DNA template, incorporating ribonucleotides one at a time. (also proofreading)
- Termination: less well understood. This event appears to be coupled to site selection for polyadenylation
Transcription Factor
- Ligand binding can “induce” their activity by permitting them to ”shuttle into” the nucleus. (“inducible” Cre recombinase)
- Combinations of TFs help to specify distinct cell types
- reprogramming: some TFs could specify cell types
- ENCODE project: evidence in support of the combinatorial association of transcription factors => cell type; developmental stage; level of expression
- Chromatin immunoprecipitation: identify putative TF-binding sites, may overestimate the number of binding sites
=======================================
- co-activators & co-repressors: should not directly bind to the DNA but should instead bind to other proteins
- co-activators: e.g. histone acetylases these enzymes add acetyl groups on to specific histone proteins
- co-repressors: e.g. histone deacetylases activities counter those of the acetylases
- There are also transcriptional repressors and co-repressors that can antagonize the function of activators and co-activators, e.g. HDAC and HAT
- Enhancer activity may be restricted to a topologically-associated domain (TAD)
- Phase separation: there are molecules (proteins and RNAs) that can self-assemble to form larger molecular complexes in support of specific functions
pioneer factor
Splicing

- The primary transcript is “capped” (m7GpppN) shortly after transcription is initiated
- the RNA hybridized with non-adjacent regions of a single DNA molecule
- In mammals, splicing is a protein-assisted process.
- “RNA can splice itself”: RNAs can be catalytic => may be an intrinsic, intramolecular RNA-mediated reaction
- Spliceosome: ~100 proteins => recognize the start(GU) and end(AG) of RNA
- Alternative splicing: Genes containing multiple exons may generate different mRNAs by the regulated inclusion or exclusion of specific exons => increases the number of proteins that can be made from one gene
Example: Nova proteins
- Nova1 & Nova2: proteins can bind to the primary RNA transcript and influence whether a particular exonic sequence will be retained or excluded
- how the location of Nova binding influenced alternative splicing: Nova1/2 double KO mice
- Nova binding in the downstream intron correlates with exon inclusion while binding to the exon correlates with exclusion
Translation
Regulation may occur via:
- general regulatory proteins that affect all mRNAs
- microRNAs
- by RNA-binding proteins
Example of Translational Control
Some mRNAs are regulated by polyadenylation (length of poly(A) tail)
- “CPEB” binds to CPE (the cytoplasmic polyadenylation element) and to the protein Maskin
- Maskin can bind to the 5’ end of the mRNA via eIF4E (eukaryotic translation initiation factor 4E), which binds to the 5’ cap. When bound to Maskin, eIF4E cannot bind to the ribosome to initiate translation
- During development, phosphorylation of CPEB leads to the dissociation of Maskin from eIF4E and polyadenylation occurs via polyA polymerase (PAP). ). Multiple molecules of PABP can now bind to this tail and recruit the translation machinery.
- Thus, regulation of poly(A) length boosts translation.
Fragile X
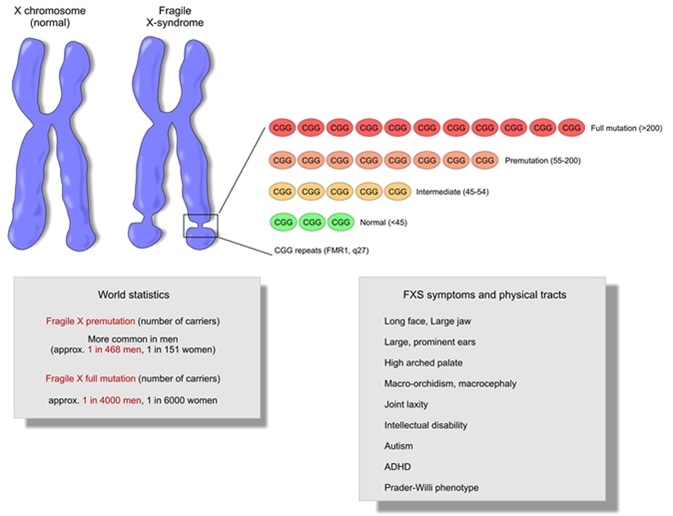
- Fragile X results from mutations in the X-linked gene, FMR1 (fragile X mental retardation-1) gene, which encodes FMRP, the fragile X mental retardation protein.
- => Fragile X is associated with a loss of FMRP expression/function
- CGG repeats are in the promoter region and not in the FMRP coding region!
- FMRP is an RNA-binding protein: represses translation by “stalling ribosomes”, i.e. preventing them from moving down the mRNAs
- Fragile X => FMRP(which represses translation) ↓ => translation of some mRNAs ↑ => encoded proteins (pre- and post-synaptic proteins) ↑ => syndrome
RNA interference & microRNAs
- RNAi: RNA molecules serve to regulate the activity of genes => reducing or “knocking down” gene expression
- MicroRNAs: endogenous small RNAs whose outcome tends to be the repression of mRNA translation
RNAi
- “antisense” nucleic acids: persumably hybirdizing to mRNAs => blocking their translation
- not only antisense, but also sens strand RNAs can supress gene expression in the nematode
- not sense strand, but ds RNAs to inhibit gene expression: most preparations of sense strand RNA were slightly contaminated with antisense RNA
- These small RNAs can “interfere” with the translation of mRNAs by the sequence-specific targeting of their destruction
Mechanism

- In the cytoplasm, ds RNAs are processed by an enzyme called “Dicer” which cuts it into small siRNAs (21-23 nts) that carry 2 NT 3’ overhangs
- The siRNAs are incorporated into the “RNA-inducing silencing complex” or “RISC”. One component, a protein called Argonaute-2 (Ago2), cleaves the target RNAs.
- It appears that only one of the 2 strands, the “guide” (antisense) strand, of the double-stranded siRNA is actually “loaded” into RISC. The other strand, the “passenger strand” is destroyed
- The guide (antisense) strand of the siRNA provides the specificity and presumably identifies its target by hybridization.
Cleavage is “catalytic” and precise
miRNAs
- Our genomes contain many “miRNAs”, which are small endogenous dsRNAs whose precursors have a hairpin-like structure
- Although some miRNAs are produced from a single transcription unit, most are produced from transcripts that make more than one miRNA
- The primary transcript is the primary miRNA (pri-mRNA), which is typically made by Pol II
- Can be processed by Dicer into siRNA-like ds RNA molecules.
- They primarily target the 3’ UT regions of mRNAs and they do not require perfect matches for their activity
- miRNAs may target hundreds of different mRNAs as the so-called “seed” or target sequence may be as short as 6 bases.
- targets shared a common, short “seed sequence” => Each miRNA appeared to target hundreds of mRNAs.
- How do miRNAs supress translation: most of the repression is accomplished by degradation of the mRNA targets
- Many miRNAs are highly evolutionarily conserved: proliferation across various spacies
Example: miR-124
- Increased levels of miR-124 led to an early reduction of progenitors, which was associated with their precocious differentiation into neurons
- Reduced levels of miR-124 expression maintained the precursors in the actively dividing state (proliferation as opposed to differentiation)
- miR-124 plays an important role in determining neuronal fate. High levels of miR-124 promotes the formation of neurons while low levels block it


留下评论